A collaborative review by ESRs Ahmad Abdullrahman from the Department of Pharmacy, Chemistry and Pharmacy Building, University of Reading (UK), and Maria Dalla Pozza from Chimie ParisTech, PSL University, CNRS, Institute of Chemistry for Life and Health Sciences (France) was published in Chemical Science in August 2022.
In this review, entitled Three’s a crowd – stabilisation, structure, and applications of DNA triplexes, the ESRs put their efforts together in proficient teamwork supervised by Dr. James Hall, Prof. Christine Cardin from the University of Reading, and Prof. Gilles Gasser from Chimie ParisTech, PSL University.
They describe the main characteristics of triplex DNA structures, focusing on their application and interaction with metal compounds, highlighting the need for additional structure characterization and biological studies.
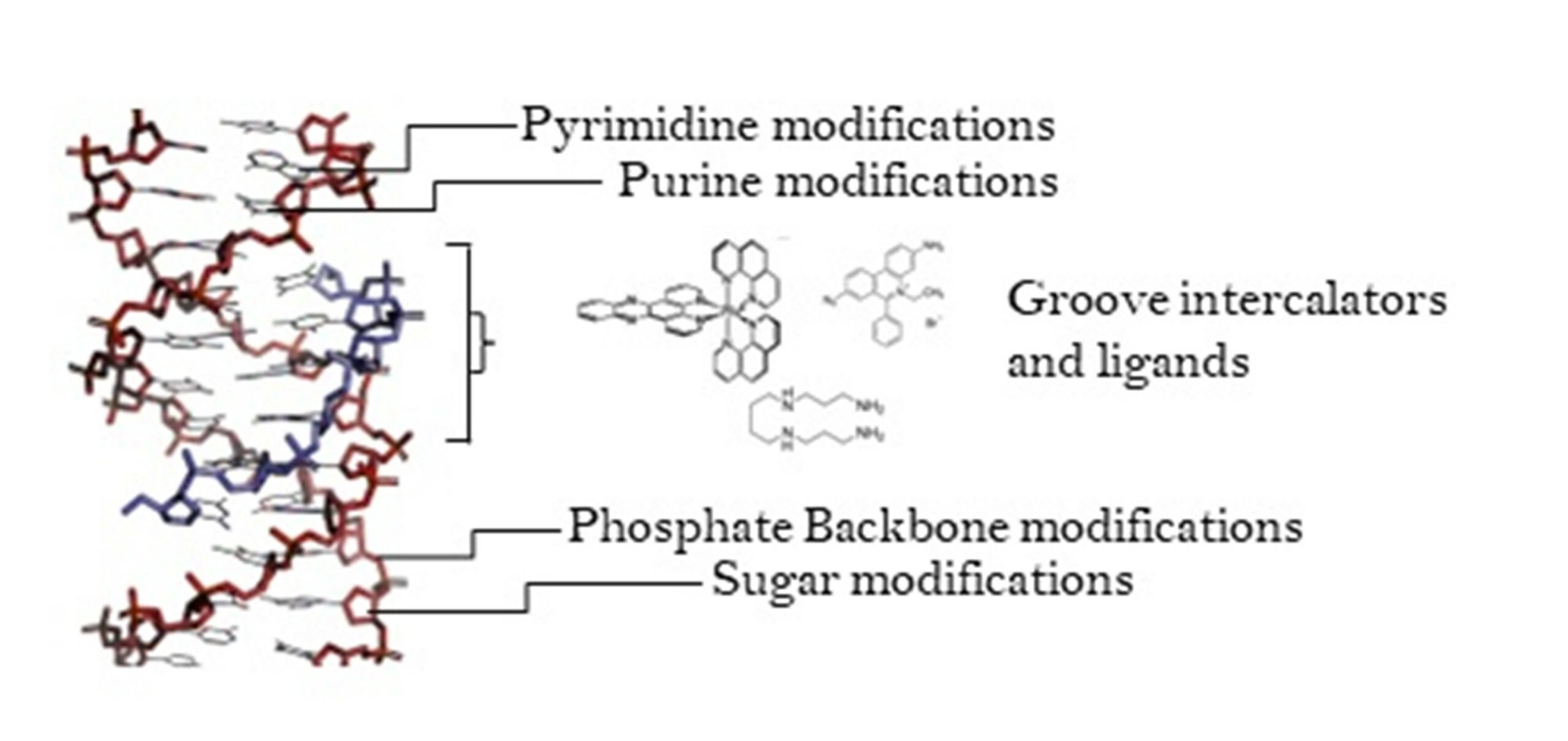
The DNA triplex may be formed naturally, during homologous recombination, or can be formed by the introduction of a synthetic triplex-forming oligonucleotide (TFO) to a DNA duplex. Among others, the most interesting feature of TFOs is their sequence specificity in binding a duplex DNA. The authors first compare the triplex structure with the canonical B-DNA structure. Subsequently, they report the main modifications at the base, sugar and phosphate backbone levels currently available to obtain a more stable structure, considering the potential in vivo conditions.
There is significant interest in developing TFOs with potential therapeutic applications, including their use as a delivery mechanism for compounds able to modify or damage DNA. However, to combine triplexes with functionalised compounds, a full understanding of triplex structure and chemical modification strategies is essential, stress the authors. In the review, these areas are discussed. Moreover, the possible use of photoactive Ruthenium polypyridyl complexes as a suitable photophysical payload for a TFO system is presented in this scientific paper. With the hope that future research will harness the peculiar characteristics of DNA triplexes.