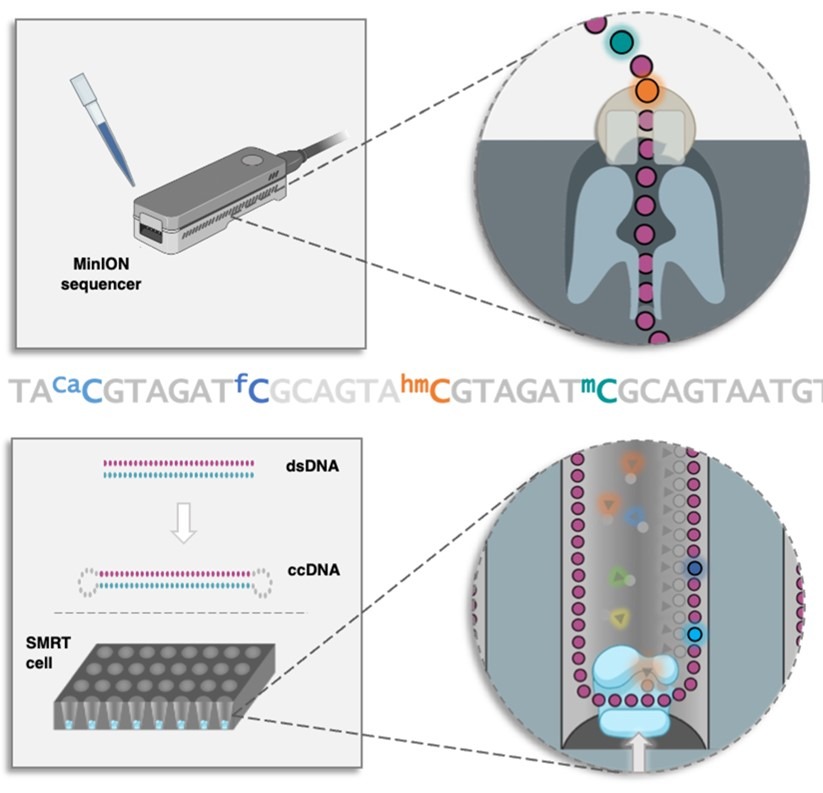
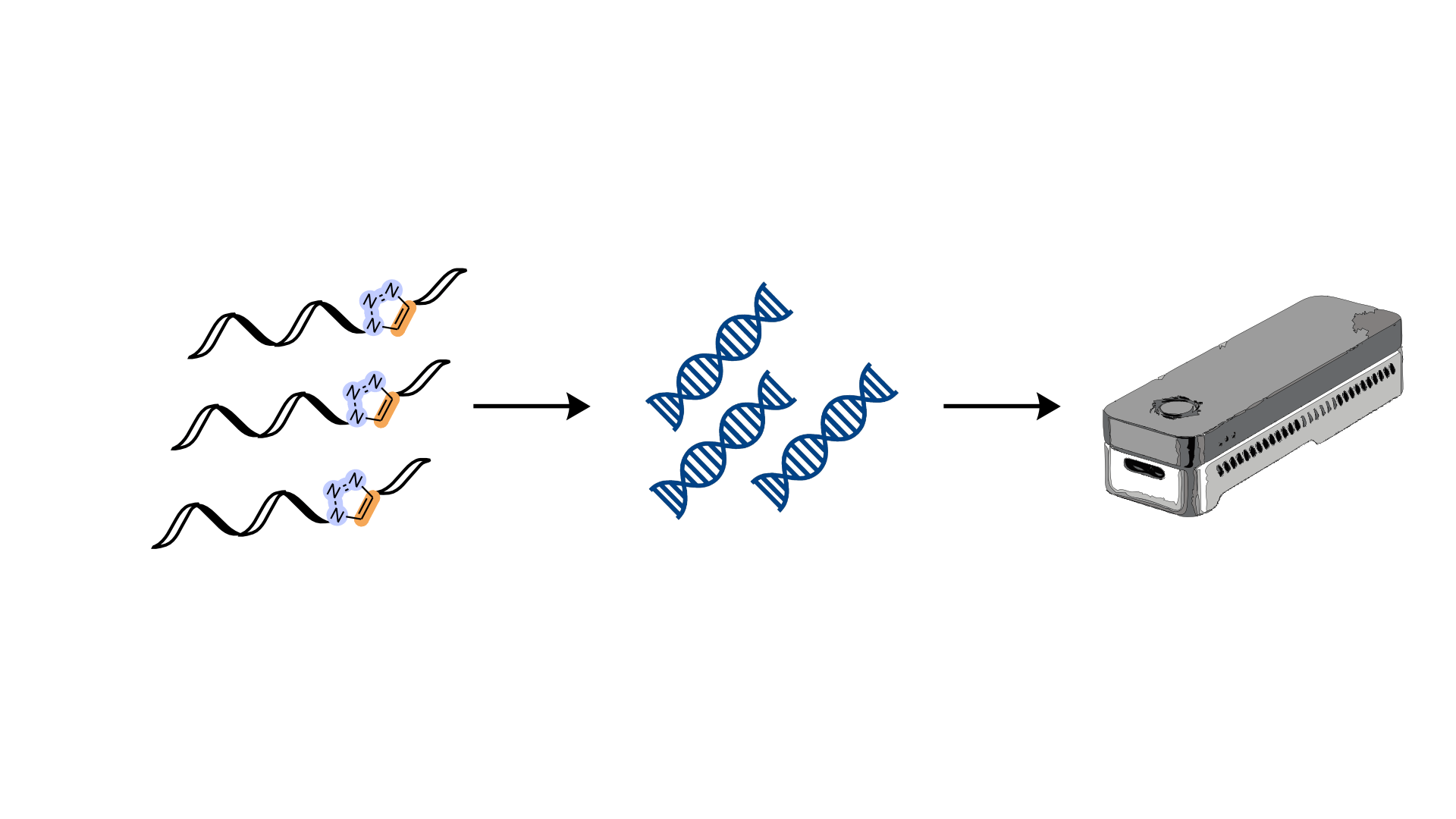
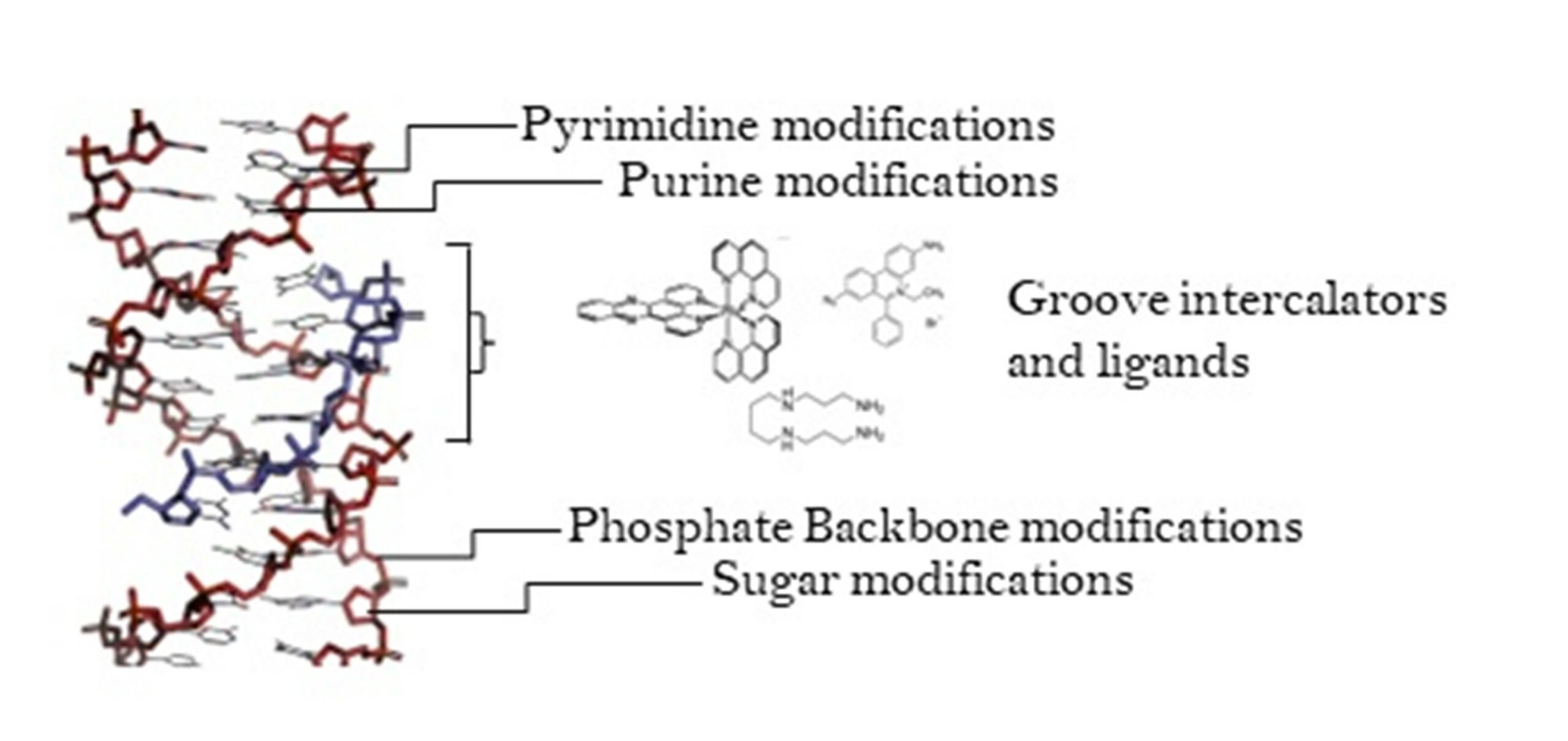
The Kellett (DCU) and Carell (LMU) groups recently published a collaborative review titled ‘Third Generation Sequencing of Epigenetic DNA’ in Angewandte Chemie. The review is open access and covers the latest developments in sequencing techniques adapted and developed for ‘third generation’ sequencing platforms, which promise to provide the fastest and most convenient means of DNA sequencing to date.
Cytosine modifications have been shown to influence gene regulation, in turn effecting disease and development, thus facile methods for sequencing these base modifications by exploiting the chemistries of these new devices is an active area of research. Despite extensive reviews covering sequencing technologies and base modifications independently, to our knowledge, this is the first publication to highlight the emerging potential of third generation sequencing technologies to expedite epigenetic research.
During the first in-person NATURE-ETN training week organised in the Institute for Chemical Epigenetic – Munich (ICE-M), Dr Markus Müller and Dr. Pascal Giehr delivered seminars focused on epigenome sequencing which provided valuable background in techniques developed to facilitate the decoding of this secondary information layer in DNA. Work Package 3 in NATURE-ETN aims to generate new techniques for sequencing and imaging epigenetic bases. DCU and LMU have access to third generation sequencing devices, so this review will provide a helpful reference point for researchers in the network.